NADH
Le nicotinamide adénine dinucléotide (NAD) is a coenzyme present in all living cells, characterized by its dinucleotide structure composed of adenosine and nicotinamide nucleotides. NAD exists in oxidized form (NAD+) and reduced form (NADH) and serves as an electron transporter in cellular redox reactions, where NAD+ acts as an oxidant and NADH as a reductant. Additionally, NAD plays a crucial role in various cellular processes, including post-translational modifications like ADP-ribosylation of newly synthesized proteins. Its synthesis involves the amino acids tryptophan and aspartate, as well as vitamin B3 (niacin), while salvage pathways recycle degraded NAD compounds. Some NAD is also converted into nicotinamide adénine dinucléotide phosphate (NADP), a coenzyme with similar biochemistry but distinct metabolic functions. The formal charge of NAD+ and NADH is -1 and -2, respectively, although the sign "+" denotes the formal charge on the nitrogen heteroatom of the nicotinamide.
Accession Number : KLM0000285 This work is released into the public domain; please see our release statement.
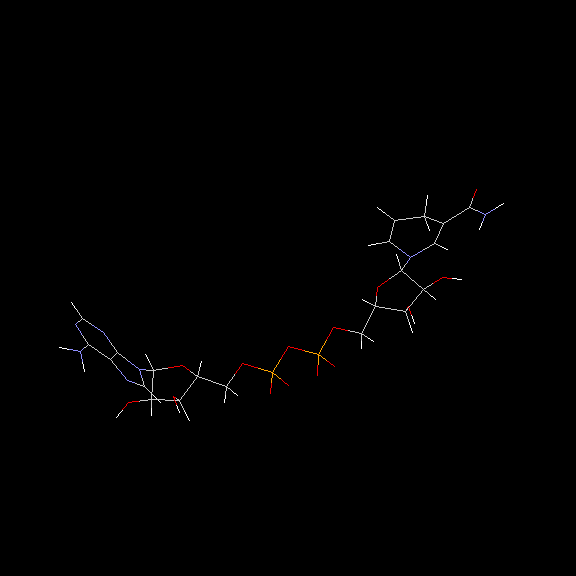
Doug Markham has contributed a molecular mechanics computation of the structure! See below for the details.
Config Rule :
% 'NADH'
config('NADH',[
substituent('dihydronicotinamide-1-yl'),
substituent('D-1-dehydroxy-5-oxy-ribofuranosyl'),
substituent('ADP-yl'),
linkage(from('ADP-yl',pho(2)),
to('D-1-dehydroxy-5-oxy-ribofuranosyl',attach_to([oxy,car(5)])),
nil,single),
linkage(from('D-1-dehydroxy-5-oxy-ribofuranosyl',car(1)),
to('dihydronicotinamide-1-yl',nit(1)),
up,single)]).
config('ADP-yl',[
substituent('D-1-dehydroxy-5-oxy-ribofuranosyl'),
substituent(diphosphopentaoxygen),
substituent(adenyl),
linkage(from('D-1-dehydroxy-5-oxy-ribofuranosyl',car(1)),
to(adenyl,nit(9)),
up,single),
linkage(from('D-1-dehydroxy-5-oxy-ribofuranosyl',attach_to([oxy,car(5)])),
to(diphosphopentaoxygen,attach_to([pho(1)])),
nil,single)]).
config('ADP-yl',[
substituent('D-1-dehydroxy-5-oxy-ribofuranosyl'),
substituent(diphosphopentaoxygen),
substituent(adenyl),
linkage(from('D-1-dehydroxy-5-oxy-ribofuranosyl',car(1)),
to(adenyl,nit(9)),
up,single),
linkage(from('D-1-dehydroxy-5-oxy-ribofuranosyl',attach_to([oxy,car(5)])),
to(diphosphopentaoxygen,attach_to([pho(1)])),
nil,single)]).
config('D-1-dehydroxy-5-oxy-ribofuranosyl',[
ring([
oxy,
anomeric(1,hyd),
car(2,hyd&&hydroxyl;),
car(3,hyd&&hydroxyl;),
car(4,oxymethyl&&hyd;)])]).
config(adenyl,[
model(adenine,[
diff(nit(9,hyd),nit(9))])]).
config(adenyl,[
model(adenine,[
diff(nit(9,hyd),nit(9))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config('D-1-dehydroxy-5-oxy-ribofuranosyl',[
ring([
oxy,
anomeric(1,hyd),
car(2,hyd&&hydroxyl;),
car(3,hyd&&hydroxyl;),
car(4,oxymethyl&&hyd;)])]).
config(adenyl,[
model(adenine,[
diff(nit(9,hyd),nit(9))])]).
config(adenyl,[
model(adenine,[
diff(nit(9,hyd),nit(9))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
config('D-1-dehydroxy-5-oxy-ribofuranosyl',[
ring([
oxy,
anomeric(1,hyd),
car(2,hyd&&hydroxyl;),
car(3,hyd&&hydroxyl;),
car(4,oxymethyl&&hyd;)])]).
config('dihydronicotinamide-1-yl',[
ring([
nit(1),
car(2,hyd)~~,
car(3,carboxamide(7)),
car(4,hyd&&hyd;),
car(5,hyd)~~,
car(6,hyd)])]).
Smiles String :
[C@2H]-1([O][C@2H]([C@2H]([OH])[C@2H]-1[OH])[C@2H2][O][P@2]([O][P@2](=[O])([O][_ C@2H2][C@1H]1([C@2H]([OH])[C@2H]([OH])[C@2H]([O]1)[N@2]1([CH]=[CH][C@2H2][C](=[_ CH]1)[C]([O-])=[NH2])))[O-])([O-])=[O])-[n]1([cH][n][c]2([c]1[n][cH][n][c]2[NH2_ ])) 'NADH'
Terminal :
% 'NADH'
c(1,12,(0,chiral))-[c(2,left)~,o(1,right)~,n(9,up)~,h(1,down)~],
c(2,12,(0,chiral))-[c(3,left)~,c(1,right)~,h(2,up)~,o(2,down)~],
c(3,12,(0,chiral))-[c(4,left)~,c(2,right)~,h(4,up)~,o(3,down)~],
c(4,12,(0,chiral))-[o(1,left)~,c(3,right)~,c(5,up)~,h(6,down)~],
c(5,12,(0,nonchiral))-[h(7,left)~,h(8,right)~,o(4,up)~,c(4,down)~],
c(6,12,(0,nonchiral))-[h(11,nil)~,n(3,flat)&,n(1,flat)&],
c(8,12,(0,nonchiral))-[n(3,flat)&,c(9,flat)&,n(9,flat)&],
c(9,12,(0,nonchiral))-[c(8,flat)&,c(10,flat)&,n(7,flat)&],
c(10,12,(0,nonchiral))-[n(10,nil)~,n(1,flat)&,c(9,flat)&],
c(12,12,(0,nonchiral))-[h(12,nil)~,n(7,flat)&,n(9,flat)&],
c(13,12,(0,chiral))-[c(14,left)~,o(10,right)~,n(11,up)~,h(13,down)~],
c(14,12,(0,chiral))-[c(15,left)~,c(13,right)~,h(14,up)~,o(11,down)~],
c(15,12,(0,chiral))-[c(16,left)~,c(14,right)~,h(16,up)~,o(12,down)~],
c(16,12,(0,chiral))-[o(10,left)~,c(15,right)~,c(17,up)~,h(18,down)~],
c(17,12,(0,nonchiral))-[o(13,left)~,h(19,right)~,h(20,up)~,c(16,down)~],
c(18,12,(0,nonchiral))-[c(19,left)~~,n(11,right)~,h(21,nil)~],
c(19,12,(0,nonchiral))-[c(20,left)~,c(18,right)~~,c(23,nil)~],
c(20,12,(0,nonchiral))-[c(21,left)~,c(19,right)~,h(24,up)~,h(25,down)~],
c(21,12,(0,nonchiral))-[c(22,left)~~,c(20,right)~,h(26,nil)~],
c(22,12,(0,nonchiral))-[n(11,left)~,c(21,right)~~,h(27,nil)~],
c(23,12,(0,nonchiral))-[o(14,nil)?,n(12,nil)#,c(19,nil)~],
h(1,1,(0,nonchiral))-[c(1,up)~],
h(2,1,(0,nonchiral))-[c(2,down)~],
h(3,1,(0,nonchiral))-[o(2,nil)~],
h(4,1,(0,nonchiral))-[c(3,down)~],
h(5,1,(0,nonchiral))-[o(3,nil)~],
h(6,1,(0,nonchiral))-[c(4,up)~],
h(7,1,(0,nonchiral))-[c(5,right)~],
h(8,1,(0,nonchiral))-[c(5,left)~],
h(9,1,(0,nonchiral))-[n(10,nil)~],
h(10,1,(0,nonchiral))-[n(10,nil)~],
h(11,1,(0,nonchiral))-[c(6,nil)~],
h(12,1,(0,nonchiral))-[c(12,nil)~],
h(13,1,(0,nonchiral))-[c(13,up)~],
h(14,1,(0,nonchiral))-[c(14,down)~],
h(15,1,(0,nonchiral))-[o(11,nil)~],
h(16,1,(0,nonchiral))-[c(15,down)~],
h(17,1,(0,nonchiral))-[o(12,nil)~],
h(18,1,(0,nonchiral))-[c(16,up)~],
h(19,1,(0,nonchiral))-[c(17,left)~],
h(20,1,(0,nonchiral))-[c(17,down)~],
h(21,1,(0,nonchiral))-[c(18,nil)~],
h(22,1,(0,nonchiral))-[n(12,nil)~],
h(23,1,(0,nonchiral))-[n(12,nil)~],
h(24,1,(0,nonchiral))-[c(20,down)~],
h(25,1,(0,nonchiral))-[c(20,up)~],
h(26,1,(0,nonchiral))-[c(21,nil)~],
h(27,1,(0,nonchiral))-[c(22,nil)~],
n(1,14,(0,nonchiral))-[c(6,flat)&,c(10,flat)&],
n(3,14,(0,nonchiral))-[c(6,flat)&,c(8,flat)&],
n(7,14,(0,nonchiral))-[c(9,flat)&,c(12,flat)&],
n(9,14,(0,nonchiral))-[c(1,down)~,c(12,flat)&,c(8,flat)&],
n(10,14,(0,nonchiral))-[h(9,nil)~,h(10,nil)~,c(10,nil)~],
n(11,14,(0,nonchiral))-[c(18,left)~,c(22,right)~,c(13,down)~],
n(12,14,(0,nonchiral))-[h(22,nil)~,h(23,nil)~,c(23,nil)#], o(1,16,(0,nonchiral))-[c(1,left)~,c(4,right)~],
o(2,16,(0,nonchiral))-[h(3,nil)~,c(2,up)~],
o(3,16,(0,nonchiral))-[h(5,nil)~,c(3,up)~],
o(4,16,(0,nonchiral))-[p(1,right)~,c(5,down)~],
o(5,16,(0,nonchiral))-[p(1,left)~,p(2,right)~],
o(6,16,(-5.0E-01,nonchiral))-[p(1,up)?],
o(7,16,(-5.0E-01,nonchiral))-[p(1,down)?],
o(8,16,(-5.0E-01,nonchiral))-[p(2,left)?],
o(9,16,(-5.0E-01,nonchiral))-[p(2,down)?],
o(10,16,(0,nonchiral))-[c(13,left)~,c(16,right)~],
o(11,16,(0,nonchiral))-[h(15,nil)~,c(14,up)~],
o(12,16,(0,nonchiral))-[h(17,nil)~,c(15,up)~],
o(13,16,(0,nonchiral))-[c(17,right)~,p(2,up)~],
o(14,16,(0,nonchiral))-[c(23,nil)?],
p(1,31,(0,nonchiral))-[o(4,left)~,o(5,right)~,o(7,up)?,o(6,down)?],
p(2,31,(0,nonchiral))-[o(5,left)~,o(8,right)?,o(9,up)?,o(13,down)~]
The Terminals for all the Config Rules are in Prolog Definite Clause Grammar (DCG) form.They can be checked in the Manual here.
The compound's PDB file can be seen here.
Doug Markham of the Institute for Cancer Research, Fox Chase Cancer Center,Philadelphia, PA, has contributed the following structure for NADH. He computed this structure in sdf format using MacroModel, a molecular mechanics program. We have used Babel to convert the .sdf format to PDB format. You'll find it interesting to compare these structures to those computed using CONCORD.
Many thanks Doug!
