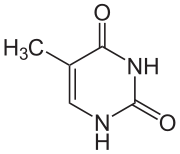
Thymine
Thymine is a pyrimidine nucleobase that pairs with adenine in DNA. It is one of the four nitrogenous bases found in DNA and RNA.
Thymine is essential for DNA replication and transcription. It also plays a role in gene regulation.
Here are some of the important functions of thymine:
- DNA replication: Thymine pairs with adenine during DNA replication. This pairing ensures that the genetic code is passed on accurately from one generation to the next.
- Transcription: Thymine is transcribed into uracil in RNA. This allows the genetic code to be read by the ribosomes and translated into proteins.
- Gene regulation: Thymine can be modified by methylation, which can affect gene expression. For example, methylation of thymine can silence genes that are associated with cancer.
Thymine deficiency is rare, but it can occur in people with certain genetic disorders. Thymine deficiency can lead to a variety of health problems, including neurological problems, immune system deficiencies, and cancer.
Accession Number : KLM0000385 This work is released into the public domain; please see our release statement.
Doug Markham has contributed a molecular mechanics computation of the structure! See below for the details.
Config Rule :
% thymine
config(thymine,[
ring([
nit(1,hyd)&,
car(2,oxy?)&,
nit(3,hyd)&,
car(4,oxy?)&,
car(5,methyl(7))&,
car(6,hyd)&])]).
config(thymine,[
ring([
nit(1,hyd)&,
car(2,oxy?)&,
nit(3,hyd)&,
car(4,oxy?)&,
car(5,methyl(7))&,
car(6,hyd)&])]).
Smiles String :
[cH]1([nH][c]([nH][c]([c]1[CH3])[O-])[O-]) thymine
Terminal :
% thymine
c(2,12,(0,nonchiral))-[o(1,nil)?,n(1,flat)&,n(3,flat)&],
c(4,12,(0,nonchiral))-[o(2,nil)?,n(3,flat)&,c(5,flat)&],
c(5,12,(0,nonchiral))-[c(7,right)~,c(4,flat)&,c(6,flat)&],
c(6,12,(0,nonchiral))-[h(6,nil)~,c(5,flat)&,n(1,flat)&],
c(7,12,(0,nonchiral))-[c(5,left)~,h(3,right)~,h(5,up)~,h(4,down)~],
h(1,1,(0,nonchiral))-[n(1,nil)~],
h(2,1,(0,nonchiral))-[n(3,nil)~],
h(3,1,(0,nonchiral))-[c(7,left)~],
h(4,1,(0,nonchiral))-[c(7,up)~],
h(5,1,(0,nonchiral))-[c(7,down)~],
h(6,1,(0,nonchiral))-[c(6,nil)~],
n(1,14,(0,nonchiral))-[h(1,nil)~,c(6,flat)&,c(2,flat)&],
n(3,14,(0,nonchiral))-[h(2,nil)~,c(2,flat)&,c(4,flat)&],
o(1,16,(0,nonchiral))-[c(2,nil)?],
o(2,16,(0,nonchiral))-[c(4,nil)?]
The Terminals for all the Config Rules are in Prolog Definite Clause Grammar (DCG) form.They can be checked in the Manual here.
The compound's PDB file can be seen here.
Doug Markham of the Institute for Cancer Research, Fox Chase Cancer Center,Philadelphia, PA, has contributed the following structure for thymine. He computed this structure in sdf format using MacroModel, a molecular mechanics program. We have used Babel to convert the .sdf format to PDB format. You'll find it interesting to compare these structures to those computed using CONCORD.
Many thanks Doug!