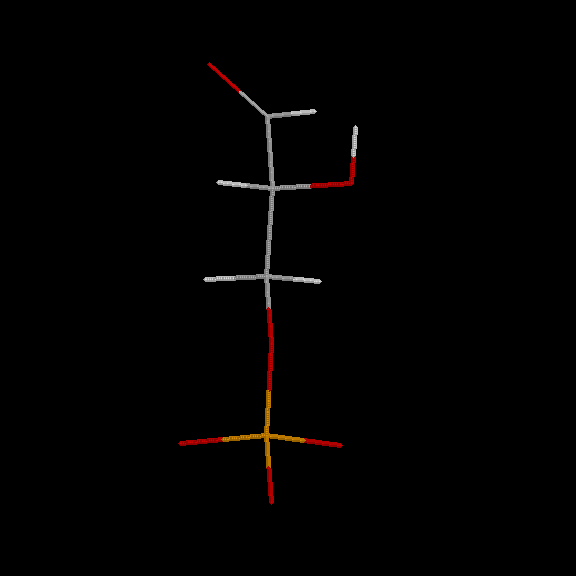
D-glyceraldehyde-3-phosphate
Glyceraldehyde-3-phosphate, also known as 3-phosphoglyceraldehyde and commonly abbreviated as G3P or GAP (or PGAL for English speakers), is an organic compound that plays a central role in several metabolic pathways in most living organisms. It is a phosphoric ester of glyceraldehyde, with only the D-glyceraldehyde-3-phosphate enantiomer being biologically active. It is an essential metabolite in glycolysis and is also involved in the non-oxidative phase of the pentose phosphate pathway.
Accession Number : KLM0000187This work is released into the public domain; please see our release statement.
Synonyms :
- glyceraldehyde-3-phosphate
- D-glyceraldehyde 3-phosphate
- G3P
Config Rule :
% 'D-glyceraldehyde-3-phosphate'
config('D-glyceraldehyde-3-phosphate',[
top(aldehyde),
center(car(2,hyd&&hydroxyl;)),
bottom(methylphosphate)]).
Smiles String :
[CH]([C@2H]([C@2H2][O][P@2](=[O])([O-])[O-])[OH])=[O] 'D-glyceraldehyde-3-phosphate'
Terminal :
% 'D-glyceraldehyde-3-phosphate'
c(1,12,(0,nonchiral))-[h(3,nil)~,o(3,nil)~~,c(2,down)~],
c(2,12,(0,chiral))-[h(1,left)~,o(1,right)~,c(1,up)~,c(3,down)~],
c(3,12,(0,nonchiral))-[h(4,left)~,h(5,right)~,c(2,up)~,o(2,down)~],
h(1,1,(0,nonchiral))-[c(2,right)~],
h(2,1,(0,nonchiral))-[o(1,nil)~],
h(3,1,(0,nonchiral))-[c(1,nil)~],
h(4,1,(0,nonchiral))-[c(3,right)~],
h(5,1,(0,nonchiral))-[c(3,left)~],
o(1,16,(0,nonchiral))-[c(2,left)~,h(2,nil)~],
o(2,16,(0,nonchiral))-[p(1,right)~,c(3,up)~],
o(3,16,(0,nonchiral))-[c(1,nil)~~],
o(4,16,(-6.666666666666666E-01,nonchiral))-[p(1,left)?],
o(5,16,(-6.666666666666666E-01,nonchiral))-[p(1,up)?],
o(6,16,(-6.666666666666666E-01,nonchiral))-[p(1,down)?],
p(1,31,(0,nonchiral))-[o(2,left)~,o(4,right)?,o(6,up)?,o(5,down)?]
The Terminals for all the Config Rules are in Prolog Definite Clause Grammar (DCG) form.They can be checked in the Manual here.
The compound's PDB file can be seen here.
Fischer Diagram :

