ADP Assay Kits
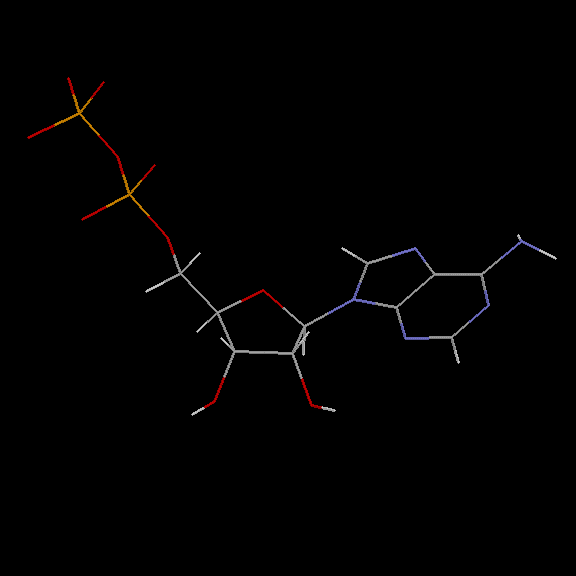
Accession Number : KLM0000039 This work is released into the public domain; please see our release statement.
Doug Markham has contributed a molecular mechanics computation of the structure! See below for the details.
Synonyms :
adenosine diphosphate
adenosine 5'-diphosphate
5' adenylate diphosphate
rADP
5' rADP
Config Rule :
% 'ADP'
Adenosine diphosphate (ADP) is a nucleoside phosphate comprised of a ribonucleoside and two phosphate groups.
Ribose as its sugar and two phosphate groups attached. Its nucleoside contains a purine base, i.e. an adenine attached to the ribose sugar. It has two phosphate groups attached to the nucleoside. The nucleoside is a pentose sugar backbone with a purine base adenine attached to it (at the 1′ carbon). The phosphate groups are bonded in series to the 5′ carbon of the pentose sugar.
Common biological reactions
ADP can be derived from adenosine triphosphate (ATP). It can be interconverted to ATP. In particular, ATP is dephosphorylated by ATPases to produce ADP. ADP in turn may be phosphorylated to become ATP. In plants, this conversion of ADP into ATP is enabled via photosynthetic pathways, as light energy is stored as chemical energy in ATP. In animals, energy can be drawn from the breakdown of foodstuff. Glucose from dietary sources, in essence, is used to gather energy via glycolysis, aerobic respiration, and fermentation. The energy is stored especially in the form of ATP. The breakdown of one phosphory bond of ATP can generate about 30.5 kilojoules per mole of ATP.1
ADP can be degraded to produce adenosine monophosphate (AMP, an adenine nucleotide with only one phosphate). Combining two molecules of ADP during ATP synthesis by the action of the enzyme adenylate kinase leads to the formation of AMP: 2 ADP → ATP + AMP.
Biological functions
ADP is essential in photosynthesis and glycolysis. It is the end-product when adenosine triphosphate ATP loses one of its phosphate groups. The energy released in the process is used to power up many vital cellular processes. ADP reconverts to ATP by the addition of a phosphate group to ADP. This occurs in processes such as substrate-level phosphorylation, oxidative phosphorylation, and photophosphorylation.
ADP is also important during the activation of platelets. It is stored inside the platelet and is released to interact with ADP receptors (e.g. P2Y1 receptors, P2Y12 receptors, etc.) on platelets.
config('ADP',[
substituent('D-1-dehydroxy-5-oxy-ribofuranosyl'),
substituent(adenyl),
substituent(diphosphoryl),
linkage(from('D-1-dehydroxy-5-oxy-ribofuranosyl',car(1)),
to(adenyl,nit(9)),
up,single),
linkage(from(diphosphoryl,pho(1)),
to('D-1-dehydroxy-5-oxy-ribofuranosyl',attach_to([oxy,car(5)])),
nil,single)]).
config('D-1-dehydroxy-5-oxy-ribofuranosyl',[
ring([
oxy,
anomeric(1,hyd),
car(2,hyd&&hydroxyl;),
car(3,hyd&&hydroxyl;),
car(4,oxymethyl&&hyd;)])]).
config(adenyl,[
model(adenine,[
diff(nit(9,hyd),nit(9))])]).
config(adenine,[
model(purine,[
diff(car(6,hyd),car(6,amine(10)))])]).
config(purine,[
ring_system([
ring([
car(6,hyd)&,
car(5)&,
car(4)&,
nit(3)&,
car(2,hyd)&,
nit(1)&]),
ring([
nit(7)&,
car(8,hyd)&,
nit(9,hyd)&,
car(4)&,
car(5)&])],
conjugate(1,pseudopos([car(4),car(5)]),2,pseudopos([car(4),car(5)]))])]).
Smiles String :
[C@2H]-1([O][C@2H]([C@2H]([OH])[C@2H]-1[OH])[C@2H2][O][P@2]([O][P@2](=[O])([O-]_
)[O-])([O-])=[O])-[n]1([cH][n][c]2([c]1[n][cH][n][c]2[NH2]))
'ADP'
Terminal :
% 'ADP'
c(1,12,(0,chiral))-[c(2,left)~,o(1,right)~,n(9,up)~,h(1,down)~],
c(2,12,(0,chiral))-[c(3,left)~,c(1,right)~,h(2,up)~,o(2,down)~],
c(3,12,(0,chiral))-[c(4,left)~,c(2,right)~,h(4,up)~,o(3,down)~],
c(4,12,(0,chiral))-[o(1,left)~,c(3,right)~,c(5,up)~,h(6,down)~],
c(5,12,(0,nonchiral))-[h(7,left)~,h(8,right)~,o(4,up)~,c(4,down)~],
c(6,12,(0,nonchiral))-[h(11,nil)~,n(3,flat)&,n(1,flat)&],
c(8,12,(0,nonchiral))-[n(3,flat)&,c(9,flat)&,n(9,flat)&],
c(9,12,(0,nonchiral))-[c(8,flat)&,c(10,flat)&,n(7,flat)&],
c(10,12,(0,nonchiral))-[n(10,nil)~,n(1,flat)&,c(9,flat)&],
c(12,12,(0,nonchiral))-[h(12,nil)~,n(7,flat)&,n(9,flat)&],
h(1,1,(0,nonchiral))-[c(1,up)~],
h(2,1,(0,nonchiral))-[c(2,down)~],
h(3,1,(0,nonchiral))-[o(2,nil)~],
h(4,1,(0,nonchiral))-[c(3,down)~],
h(5,1,(0,nonchiral))-[o(3,nil)~],
h(6,1,(0,nonchiral))-[c(4,up)~],
h(7,1,(0,nonchiral))-[c(5,right)~],
h(8,1,(0,nonchiral))-[c(5,left)~],
h(9,1,(0,nonchiral))-[n(10,nil)~],
h(10,1,(0,nonchiral))-[n(10,nil)~],
h(11,1,(0,nonchiral))-[c(6,nil)~],
h(12,1,(0,nonchiral))-[c(12,nil)~],
n(1,14,(0,nonchiral))-[c(6,flat)&,c(10,flat)&],
n(3,14,(0,nonchiral))-[c(6,flat)&,c(8,flat)&],
n(7,14,(0,nonchiral))-[c(9,flat)&,c(12,flat)&],
n(9,14,(0,nonchiral))-[c(1,down)~,c(12,flat)&,c(8,flat)&],
n(10,14,(0,nonchiral))-[h(9,nil)~,h(10,nil)~,c(10,nil)~],
o(1,16,(0,nonchiral))-[c(1,left)~,c(4,right)~],
o(2,16,(0,nonchiral))-[h(3,nil)~,c(2,up)~],
o(3,16,(0,nonchiral))-[h(5,nil)~,c(3,up)~],
o(4,16,(0,nonchiral))-[p(1,right)~,c(5,down)~],
o(5,16,(0,nonchiral))-[p(1,left)~,p(2,right)~],
o(6,16,(-5.0E-01,nonchiral))-[p(1,up)?],
o(7,16,(-5.0E-01,nonchiral))-[p(1,down)?],
o(8,16,(-6.666666666666666E-01,nonchiral))-[p(2,left)?],
o(9,16,(-6.666666666666666E-01,nonchiral))-[p(2,up)?],
o(10,16,(-6.666666666666666E-01,nonchiral))-[p(2,down)?],
p(1,31,(0,nonchiral))-[o(4,left)~,o(5,right)~,o(7,up)?,o(6,down)?],
p(2,31,(0,nonchiral))-[o(5,left)~,o(8,right)?,o(10,up)?,o(9,down)?]
The Terminals for all the Config Rules are in Prolog Definite Clause Grammar (DCG) form.They can be checked in the Manual here.
The compound's PDB file can be seen here.
Doug Markham of the Institute for Cancer Research, Fox Chase Cancer Center,Philadelphia, PA, has contributed the following structure for ADP. He computed this structure in sdf format using MacroModel, a molecular mechanics program. We have used Babel to convert the .sdf format to PDB format. You'll find it interesting to compare these structures to those computed using CONCORD.
Many thanks Doug!
